Molecular Beacon Design
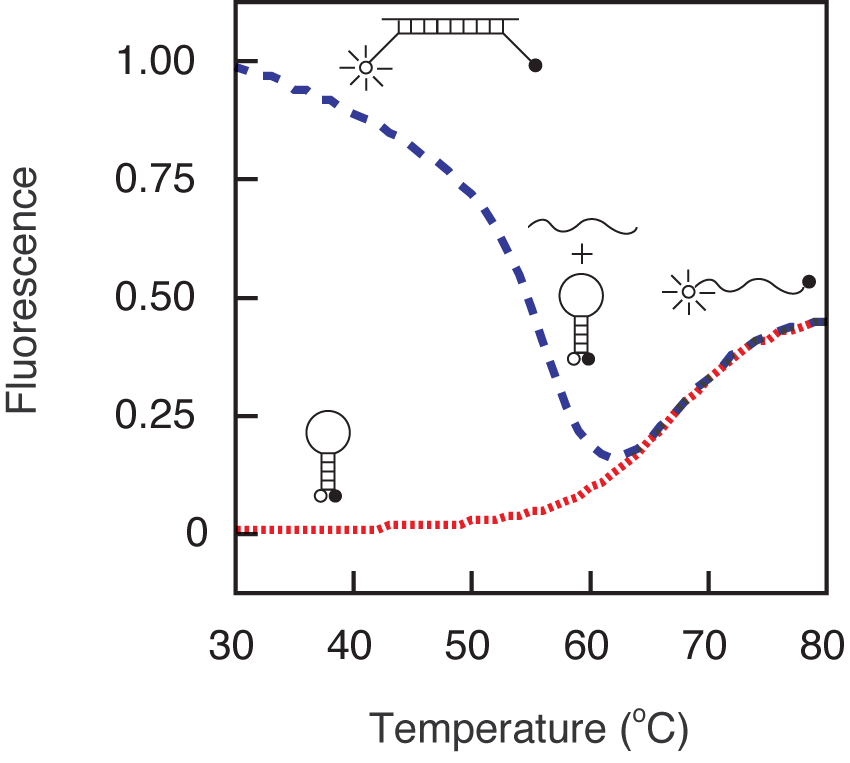
In order to design molecular beacons that function optimally under a given set of assay conditions, it is important to understand how their fluorescence changes with temperature in the presence and in the absence of their targets. As shown by the green fluorescence versus temperature trace below, at lower temperatures molecular beacons exist in a closed state, the fluorophore and the quencher are held in close proximity to each other by the hairpin stem, and there is no fluorescence. However, at high temperatures the helical order of the stem gives way to a random-coil configuration, separating the fluorophore from the quencher and restoring fluorescence. The temperature at which the stem melts depends upon the GC content and the length of the stem sequence. If a target is added to a solution containing a molecular beacon at temperatures below the melting temperature of its stem, the molecular beacon spontaneously binds to its target, dissociating the stem, and turning on its fluorescence. How the fluorescence of the probe-target hybrid varies with the temperature is indicated by the red fluorescence versus temperature trace. At low temperatures, the probe-target hybrid remains brightly fluorescent, but as the temperature is raised the probe dissociates from the target and tends to return to its hairpin state, diminishing the fluorescence significantly. The temperature at which the probe-target hybrid melts apart depends upon the GC content and the length of the probe sequence. The longer the probe and the higher its GC content, the higher the melting temperature of the probe-target hybrid. It is important to note that the probe-target hybrid melting temperature can be adjusted independently from the melting temperature of the stem by selecting a target region of appropriate length. The fluorescence versus temperature profiles of the molecular beacon that were used in this example indicate that the molecular beacon is suitable for assays that are performed below 55 ˚C, because below 55 ˚C the free molecular beacons remain dark, yet the probe-target hybrids form spontaneously and are stable.
The process of molecular beacon design begins with the selection of the probe sequence. If you are designing molecular beacons to detect the synthesis of products during polymerase chain reactions, you can select any region within the amplicon that is outside the primer binding sites. The probe sequence of the molecular beacon should be so long that at the annealing temperature of the PCR it is able to bind to its target. In order to discriminate between amplicons that differ from one another by as little as a single nucleotide substitution, the length of the probe sequence should be such that it dissociates from its target at temperatures 7-10 ˚C higher than the annealing temperature of the PCR. If, on the other hand, single-nucleotide allele discrimination is not desired, longer and more stable probes can be chosen. The melting temperature of the probe-target hybrid can be predicted using the 'percent-GC' rule or 'nearest neighbor' rules (available in most probe or primer design software packages). The prediction should be made for the probe sequence alone before choosing the stem sequences. In practice, the length of the probe sequence usually falls in the range between 15 and 30 nucleotides.
After selecting the probe sequence, two complementary arm sequences are added on either side of the probe sequence. The length and the GC content of the stem sequence is designed in such a way that at the annealing temperature of the PCR, and in the absence of the target, the molecular beacons remain closed and non-fluorescent. This is ensured by choosing a stem that melts 7-10 ˚C higher than the annealing temperature of the PCR. Usually the stems are 5-7 basepairs long and have a very high GC content (75 to 100 percent). The melting temperature of the stem can not be predicted by the percent-GC rule, since the stem is created by intramolecular hybridization. Instead, a DNA folding program, such as the IDT Oligo Analyzer, which is available on the internet at www.idtdna.com, is utilized to estimate the melting temperature of the stem. In general, 5 basepair-long GC-rich stems will melt between 55 and 60 ˚C, 6 basepair-long GC-rich stems will melt between 60 and 65 ˚C, and 7-basepair long GC-rich stems will melt between 65 and 70 ˚C. Although any arbitrary sequence can be used in designing the stems, don't use guanosine residues near the end to which the fluorophore is attached (instead, use them at the end where the quencher is attached), as guanosine residues tend to quench the fluorophore. Longer stems can be used to enhance the specificity of the molecular beacons.
It is important that the conformation assumed by the free molecular beacons be the intended hairpin structure, rather than other structures that either do not place the fluorophore in the immediate vicinity of the quencher, or that form longer stems than intended. The former will cause high background signals, and the latter will make the molecular beacons sluggish in binding to the targets. A folding of the selected sequence by the Zuker DNA folding program will reveal such problems. If unexpected secondary structures result from the choice of the stem sequence, a different stem sequence can be chosen. If, on the other hand, unexpected secondary structures arise from the identity of the probe sequence, the frame of the probe can be moved along the target sequence to obtain a probe sequence that is not self-complementary. Small stems within the probe's hairpin loop that are 2- to 3-nucleotides long do not adversely effect the performance of molecular beacons.
As with PCR primers, the sequence of the molecular beacon should be compared with the sequences of the primers, using a primer design software program to make sure that there are no regions of substantial complementarity that may cause the molecular beacon to bind to one of the primers, causing primer extension. Also, the primers that are used should be designed to produce a relatively short amplicon. In general, the amplicons should be less than 150-basepairs long. Molecular beacons are internal probes that must compete with the other strand of the amplicon for binding to the strand that contains their target sequence. Having a shorter amplicon allows the molecular beacons to compete more efficiently, and therefore produces stronger fluorescence signals during real-time PCR. In addition, smaller amplicons result in more efficient amplification. And finally, the magnitude of the molecular beacon signal can be increased by performing asymmetric PCR, in which the primer that makes the strand that is complementary to the molecular beacon is present at a slightly higher concentration than the other primer.
Software packages, such as Beacon DesignerTM developed by PREMIER Biosoft International (www.premierbiosoft.com), can be used to design efficient molecular beacon probes. The module was developed with our consultation to ensure that it adheres to the guidelines laid. The program designs efficient primers and probes by avoiding homologies and template structures.
In addition, companies, such as NYtor (www.nytor.nl), are specialized in designing real-time PCR assays that utilize molecular beacons for the detection of single nucleotide polymorphisms and high-throughput multiplex diagnostic assays.
A detailed description on the design, synthesis and application of molecular beacons used for genotyping single nucleotide polymorphisms appeared in:
-
Marras SAE, Kramer FR, and Tyagi S (2003) Genotyping single nucleotide polymorphisms
with molecular beacons. In Kwok, P. Y. (ed.), Single nucleotide
polymorphisms: methods and protocols. The Humana Press Inc., Totowa, NJ, Vol. 212, pp. 111-128.
- Vet, J.A.M. and Marras, S.A.E. (2004) Design and optimization of molecular beacon real-time polymerase chain reaction assays.
In Herdewijn, P. (ed.), Oligonucleotide synthesis: Methods and Applications.
Humana Press, Totowa, NJ, Vol. 288, pp. 273-290.
Recent Publications from our group
Banada PP, Green R, Streck D, Kurathi R, Reiss R, Banik S, Montalvan I, Jones R, Marras SAE, Chakravorty S, and Alland D (2023) An expanded RT-PCR melting temperature coding assay to rapidly identify all known SARS-CoV-2 variants and sub-variants of concern. Scientific Reports 13. 21927. PMID: 38081834: PubMed Link
Ebraham L, Xu C, Wang A, Hernandez C, Siclari N, Rajah D, Walter L, Marras SAE, Tyagi S, Fine DH, Daep CA, and Chang TL (2023) Oral Epithelial cells expressing low or undetectable levels of human angiotensin-converting enzyme 2 are susceptible to SARS-CoV-2 virus infection in vitro. Pathogens 12. PMID: 37375533: PubMed Link